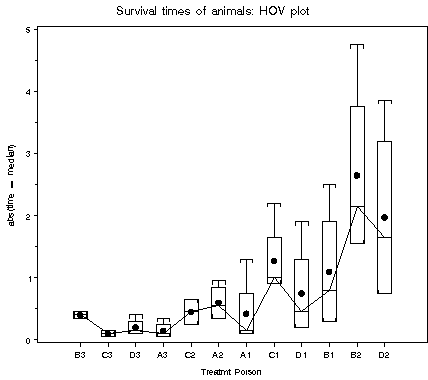
| hovplot | Boxplot display of homogeneity of variance tests | hovplot |
The HOVPLOT macro provides a graphical display of information related to the Levine and Brown-Forsythe tests of homogeneity of variance in factorial ANOVA designs.
Of the recommended tests of homogeneity of variance, the Levine test and the Brown-Forsythe test take simple forms amenable to graphical display. Both of these are based on an ANOVA of simple functions of a dispersion variable,
Z = abs (Y - median) [Brown-Forsythe] Z = abs (Y - mean) [Levine, C<TYPE=ABS>] Z = (Y - mean)^2 [Levine, C<TYPE=SQUARE>]
O'Brien's (1979) test, is a modification of Levine's Z^2, with a more complex formula. The Brown-Forsythe test appears to have the greatest power for detecting non-constant variance.
The HOVPLOT macro displays these quantities by a set of boxplots, one for each cell in the design. Lack of homogeneity of variance is indicated by differences in spread across cells.
Statistical tests are provided by PROC GLM using the
HOVTEST= option on the MEANS statement, but only for one-way
designs. The HOVPLOT macro extends this test to n-way designs by
combining multiple CLASS= variables into a single combined variable
(whose values should be distinct) representing all the cells in
the design.
The HOVPLOT macro is defined with keyword parameters. The VAR= and
CLASS= parameters are required.
The arguments may be listed within parentheses in any order, separated
by commas. For example:
%hovplot(data=animals, var=time, class=Treatmt Poison, sortby=_iqr_);
DATA=_LAST_]
variable(s)
CLASS= variable(s)
CONNECT=1]
BF or LEVINE, LEVINE(TYPE=ABS),
LEVINE(TYPE=SQUARE), or OBRIEN. Use METHOD= (null) or
METHOD=NONE to suppress the numerical test.
[Default: METHOD=BF]
CENTER=MEDIAN]
FUNCTION=ABS]
NAME=HOVPLOT]
GOUT=GSEG]
data animals;
do poison=1 to 3;
do rep = 1 to 4;
do treatmt='A', 'B', 'C', 'D';
group = treatmt || put(poison,1.);
input time @;
time = time*10;
output;
end;
end;
end;
label treatmt='Treatment' time='Survival time (hrs)'
poison='Poison';
datalines;
0.31 0.82 0.43 0.45
0.45 1.10 0.45 0.71
0.46 0.88 0.63 0.66
0.43 0.72 0.76 0.62
0.36 0.92 0.44 0.56
0.29 0.61 0.35 1.02
0.40 0.49 0.31 0.71
0.23 1.24 0.40 0.38
0.22 0.30 0.23 0.30
0.21 0.37 0.25 0.36
0.18 0.38 0.24 0.31
0.23 0.29 0.22 0.33
%include macros(hovplot); *-- or include in an autocall library; title 'Survival times of animals: HOV plot'; %hovplot(data=animals, var=time, class=Treatmt Poison, sortby=_iqr_);Output