| sprdplot | Find power transformations to equalize variance | sprdplot |
The spread-level plot has the property that *if* the relationship between log10(Interquartile range) and log10(Median) is reasonably linear, then the recommended power is p = 1 - slope, and the transformation is
/ y**p, p > 0
y --> | log(p), p = 0
\ -100y**p, p < 0
The macro chooses the best power(s) from a list of simple integers
and half-integers (PLIST=), and creates new variables using those
transformations.
%sprdplot(data=animals, var=survive, class=treat poison);
DATA=_LAST_]
abs(minimum) value, to ensure that
all values are positive.
BEST=1, the transformed variable is named T_&var. If BEST>1, the variables
are named T_1&var, T_2&var, ... [Default: PREFIX=T_]
&var [Default: BEST=1]
PPLOT=N]
GPLOT=Y]
HTEXT=1.7]
%include macros(sprdplot); *-- or include in an autocall library;
title 'Survival times of animals';
* Hand etal #403, from Box & Cox;
data animals;
do poison=1 to 3;
do rep = 1 to 4;
do treatmt='A', 'B', 'C', 'D';
input time @;
time = time*10;
output;
end;
end;
end;
label treatmt='Treatment' time='Survival time (hrs)';
cards;
0.31 0.82 0.43 0.45 0.45 1.10 0.45 0.71
0.46 0.88 0.63 0.66 0.43 0.72 0.76 0.62
0.36 0.92 0.44 0.56 0.29 0.61 0.35 1.02
0.40 0.49 0.31 0.71 0.23 1.24 0.40 0.38
0.22 0.30 0.23 0.30 0.21 0.37 0.25 0.36
0.18 0.38 0.24 0.31 0.23 0.29 0.22 0.33
;
*-- Check for variance dependent on mean;
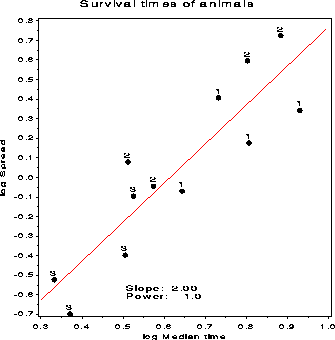
%sprdplot(data=animals, class=poison treatmt, var=time);
This produces the graph, chooses p= -1 (i.e., -100/Time), which is
saved in the variable T_TIME.
*-- Analyze the transformed response (T_TIME = -1/Time); proc glm data=animals; class poison treatmt; model t_time = poison | treatmt;